Srinithi Muthurman
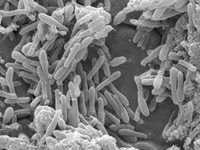
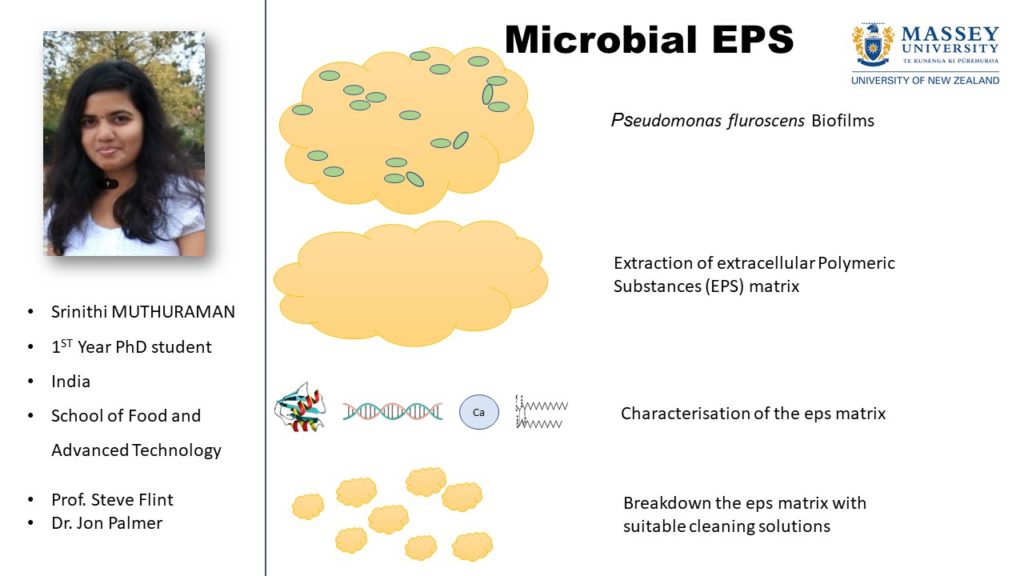
We know that biofilms are comprised of an extracellular matrix containing mixtures of polysaccharides, lipids, extracellular DNA and proteins. We hypothesise that there is much more to the composition of the matrix. To test this hypothesis will require a new approach to the study of EPS that will involve bio-chemical analysis and modelling. Our understanding of the composition and complexity of the extracellular matrix in biofilms continues to evolve. For example, two distinct functional roles for the Bacillus amphipathic protein that contributes to the hydrophobicity of B. subtilis are now known. This protein exists as a monomer or dimer depending on the redox potential in the biofilm and this has implications in the architecture of the biofilm. Another example of a recent development in knowledge of EPS is confirmation that the release of extracellular DNA into the matrix is an active process associated with DNABII proteins, independent of cell lysis and occurs in a specific location in the bacterial outer membrane.
In a study of biofilms in dairy wastewater treatment systems a mixed population of bacteria was associated with a tissue-like, highly structured extracellular mass containing poly-β hydroxyl alkanoate (PHA), a polyester produced by microorganisms used as an energy source. This exogenous rather than endogenous presence of PHA was unexpected although another report appeared recently involving PHA in biofilm formation. Questions that we would like to answer are: how do these microbial compounds become involved in the extracellular matrix?, what role do they have in the survival of the cells comprising the biofilm?, how are they involved in generating the EPS structure?, what other molecules in the extracellular matrix are involved?, how do these substances vary in different conditions?, how do they contribute to the re-colonisation of surfaces with microorganisms (important in the food industry) and finally can we model the production of EPS to develop strategies to control EPS production?
To answer these questions, metagenomics within EPS samples from different environments will scope out potential candidate microflora for EPS production. The chemical composition of the EPS taken will be determined with GC-MS analysis and the structure will be determined through NMR7. We will model the conditions leading to EPS production using a one dimensional (N1) model as such models can input a number of factors such as thickness and concentration of different components. Predominant microflora will be isolated and RNA-seq analysis will be used to investigate gene expression under conditions identified by the model as being critical for EPS production.
Supervisors
Steve Flint
Jon Palmer